Abstract
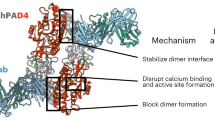
Peptidylarginine deiminase 4 (PAD4) is a Ca2+-dependent enzyme that catalyzes the conversion of protein arginine residues to citrulline. Its gene is a susceptibility locus for rheumatoid arthritis. Here we present the crystal structure of Ca2+-free wild-type PAD4, which shows that the polypeptide chain adopts an elongated fold in which the N-terminal domain forms two immunoglobulin-like subdomains, and the C-terminal domain forms an α/β propeller structure. Five Ca2+-binding sites, none of which adopt an EF-hand motif, were identified in the structure of a Ca2+-bound inactive mutant with and without bound substrate. These structural data indicate that Ca2+ binding induces conformational changes that generate the active site cleft. Our findings identify a novel mechanism for enzyme activation by Ca2+ ions, and are important for understanding the mechanism of protein citrullination and for developing PAD-inhibiting drugs for the treatment of rheumatoid arthritis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Vossenaar, E.R., Zendman, A.J.W., Venrooij, W.J. & Pruijn, G.J.M. PAD, a growing family of citrullinating enzymes: genes, features and involvement in disease. BioEssays 25, 1106–1118 (2003).
Imparl, J.M., Senshu, T. & Graves, D.J.M. Studies of calcineurin-calmodulin interaction: probing the role of arginine residues using peptidylarginine deiminase. Arch. Biochem. Biophys. 318, 370–377 (1995).
Lamensa, J.W. & Moscarello, M.A. Deimination of human myelin basic protein by a peptidylarginine deiminase from bovine brain. J. Neurochem. 61, 987–996 (1993).
Tarcsa, E. et al. Protein unfolding by peptidylarginine deiminase. J. Biol. Chem. 271, 30709–30716 (1996).
Watanabe, K. et al. Combined biochemical and immunochemical comparison of peptidylarginine deiminases present in various tissues. Biochim. Biophys. Acta 966, 375–383 (1988).
Terakawa, H., Takahara, H. & Sugawara, K. Three types of mouse peptidylarginine deiminase: characterization and tissue distribution. J. Biochem. 110, 661–666 (1991).
Guerrin M. et al. cDNA cloning, gene organization and expression analysis of human peptidylarginine deiminase type I. Biochem. J. 370, 167–174 (2003).
Ishigami, A. et al. Human peptidylarginine deiminase type II: molecular cloning, gene organization, and expression in human skin. Arch. Biochem. Biophys. 407, 25–31 (2002).
Kanno, T. et al. T. Human peptidylarginine deiminase type III: molecular cloning and nucleotide sequence of the cDNA, properties of the recombinant enzyme, and immunohistochemical localization in human skin. J. Invest. Dermatol. 115, 813–823 (2000).
Nakashima, K. et al. Molecular characterization of peptidylarginine deiminase in HL-60 cells induced by retinoic acid and 1α, 25-dihydroxyvitamin D3 . J. Biol. Chem. 274, 27786–27792 (1999).
Chavanas, S. et al. Comparative analysis of the mouse and human peptidylarginine deiminase gene clusters reveals highly conserved non-coding segments and a new human gene, PADI6. Gene 330, 19–27 (2004).
Senshu, T., Akiyama, K., Ishigami, A. & Nomura, K. Studies on specificity of peptidylarginine deiminase reactions using an immunochemical probe that recognizes an enzymatically deiminated partial sequence of mouse keratin K1. J. Dermatol. Sci. 21, 113–126 (1999).
Ishida-Yamamoto, A. et al. Sequential reorganization of cornified cell keratin filaments involving filaggrin-mediated compaction and keratin 1 deimination. J. Invest. Dermatol. 118, 282–287 (2002).
Moscarello, M.A., Pritzker, L.B., Mastronardi, F.G. & Wood, D.D. Peptidylarginine deiminase: a candidate factor in demyelinating disease. J. Neurochem. 81, 335–343 (2002).
Pritzker, L.B., Nguyen, T.A. & Moscarello, M.A. The developmental expression and activity of peptidylarginine deiminase in the mouse. Neurosci. Lett. 266, 161–164 (1999).
Rogers, G., Winter, B., McLaughlan, C., Powell, B. & Nesci, T. Peptidylarginine deiminase of the hair follicle: characterization, localization, and function in keratinizing tissues. J. Invest. Dermatol. 108, 700–707 (1997).
Hagiwara, T., Nakashima, K., Hirano, H., Senshu, T. & Yamada, M. Deimination of arginine residues in nucleophosmin/B23 and histones in HL-60 granulocytes. Biochem. Biophys. Res. Commun. 290, 979–983 (2002).
Nakashima, K., Hagiwara, T. & Yamada, M. Nuclear localization of peptidylarginine deiminase V and histone deimination in granulocytes. J. Biol. Chem. 277, 49562–49568 (2002).
Wright, P.W. et al. ePAD, an oocyte and early embryo-abundant peptidylarginine deiminase-like protein that localizes to egg cytoplasmic sheets. Dev. Biol. 256, 74–89 (2003).
Ishida-Yamamoto, A. et al. Decreased deiminated keratin K1 in psoriatic hyperproliferative epidermis. J. Invest. Dermatol. 114, 701–705 (2000).
Wood, D.D., Bilbao, J.M., O'Connors, P. & Moscarello, M.A. Acute multiple sclerosis (Marburg type) is associated with developmentally immature myelin basic protein. Ann. Neurol. 40, 18–24 (1996).
Masson-Bessière, C. et al. The major synovial targets of the rheumatoid arthritis-specific antifilaggrin autoantibodies are deiminated forms of the α- and β-chains of fibrin. J. Immunol. 166, 4177–4184 (2001).
van Boekel, M.A., Vossenaar, E.R., van den Hoogen, F.H. & van Venrooij, W.J. Autoantibody systems in rheumatoid arthritis: specificity, sensitivity and diagnostic value. Arthritis Res. 4, 87–93 (2002).
Suzuki, A. et al. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat. Genet. 34, 395–402 (2003).
Humm, A., Fritsche, E., Steinbacher, S. & Huber, R. Crystal structure and mechanism of human L-arginine: glycine amidinotransferase: a mitochondrial enzyme involved in creatine biosynthesis. EMBO J. 16, 3373–3385 (1997).
Takahara, H., Okamoto, H. & Sugawara, K. Calcium-dependent properties of peptidylarginine deiminase from rabbit skeletal muscle. Agric. Biol. Chem. 50, 2899–2904 (1986).
Holm, L. & Sander, C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233, 123–138 (1993).
Wu, H. et al. Kinetic and structural analysis of mutant CD4 receptors that are defective in HIV gp120 binding. Proc. Natl. Acad. Sci. USA. 93, 15030–15035 (1996).
Xiong, J.P. et al. Crystal structure of the extracellular segment of integrin αVβ3. Science 294, 339–345 (2001).
Tan, K. et al. The structure of immunoglobulin superfamily domains 1 and 2 of MAdCAM-1 reveals novel features important for integrin recognition. Structure 6, 793–801 (1998).
Wonhwa, C. Membrane targeting by C1 and C2 domains. J. Biol. Chem. 276, 32407–32410 (2001).
Conti, E., Uy, M., Leighton, L., Blobel, G. & Kuriyan, J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin α. Cell 94, 193–204 (1998).
Murray-Rust, J. et al. Structural insights into the hydrolysis of cellular nitric oxide synthase inhibitors by dimethylarginine dimethylaminohydrolase. Nat. Struct. Biol. 8, 679–683 (2001).
Galkin, A. et al. Structural insight into arginine degradation by arginine deiminase, an antibacterial and parasite drug target. J. Biol. Chem. 279, 14001–14008 (2004).
Das, K. et al. Crystal structures of arginine deiminase with covalent reaction intermediates: implications for catalytic mechanism. Structure 12, 657–667 (2004).
Khorchid, A. & Ikura, M. How calpain is activated by calcium. Nat. Struct. Biol. 9, 239–241 (2002).
Ahvazi, B., Kim, H.C., Kee, S.H., Nemes, Z. & Steinert, P.M. Three-dimensional structure of the human transglutaminase 3 enzyme: binding of calcium ions changes structure for activation. EMBO J. 21, 2055–2067 (2002).
Nomura, K. Specificity and Mode of action of the muscle-type protein-arginine deiminase. Arch. Biochem. Biophys. 293, 362–369 (1991).
Jones, S. & Thornton, J.M. Protein-protein interactions: a review of protein dimer structures. Prog. Biophys. Mol. Biol. 63, 31–59 (1995).
Svergun, D.I. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 76, 2879–2866 (1999).
Arita, K., Hashimoto, H., Shimizu, T., Yamada, M. & Sato, M. Crystallization and preliminary X-ray crystallographic analysis of human peptidylarginine deiminase V. Acta Crystallogr. D 59, 2332–2333 (2003).
Pflugrath, J.W. The finer things in X-ray diffraction data collection. Acta Crystallogr. D 55, 1718–1725 (1999).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Terwilliger, T.C. & Berendzen, J. Automated MAD and MIR structure solution. Acta Crystallogr. D 55, 849–861 (1999).
de la Fortelle, E. & Bricogne, G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction method. Methods Enzymol. 276, 472–494 (1997).
Abrahams, J.P. & Leslie, A.G.W. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D 52, 30–42 (1996).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Brünger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997).
Nicholls, A., Sharp, K.A. & Honig, B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11, 281–296 (1991).
Acknowledgements
The authors thank M.Y., T.K. and K.M. for data collection at SPring-8 and N.M., N.I., M.S. and S.W. for data collection at PF-AR. This work was supported by grants-in-aid for young scientists (B) from the Japan Society of the Promotion of Science (JSPS) to H.H. (14780515), grants-in-aid for scientific research (C) from the JSPS to T.S. (15570101) and M.Y. (15570122), and by the Japan Ministry of Education, Culture, Sports, Science and Technology Project on Protein Structural and Functional Analyses.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Multiple sequence alignment of human PADs. (PDF 731 kb)
Rights and permissions
About this article
Cite this article
Arita, K., Hashimoto, H., Shimizu, T. et al. Structural basis for Ca2+-induced activation of human PAD4. Nat Struct Mol Biol 11, 777–783 (2004). https://doi.org/10.1038/nsmb799
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb799
This article is cited by
-
Bioprocessing of Epothilone B from Aspergillus fumigatus under solid state fermentation: Antiproliferative activity, tubulin polymerization and cell cycle analysis
BMC Microbiology (2024)
-
Role and intervention of PAD4 in NETs in acute respiratory distress syndrome
Respiratory Research (2024)
-
Modes of PADing
Nature Chemical Biology (2024)
-
Essential role of local antibody distribution in mediating bone-resorbing effects
Scientific Reports (2024)
-
Protein citrullination and NET formation do not contribute to the pathology of A20/TNFAIP3 mutant mice
Scientific Reports (2023)