Abstract
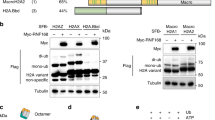
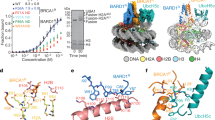
53BP1 (also called TP53BP1) is a chromatin-associated factor that promotes immunoglobulin class switching and DNA double-strand-break (DSB) repair by non-homologous end joining. To accomplish its function in DNA repair, 53BP1 accumulates at DSB sites downstream of the RNF168 ubiquitin ligase. How ubiquitin recruits 53BP1 to break sites remains unknown as its relocalization involves recognition of histone H4 Lys 20 (H4K20) methylation by its Tudor domain. Here we elucidate how vertebrate 53BP1 is recruited to the chromatin that flanks DSB sites. We show that 53BP1 recognizes mononucleosomes containing dimethylated H4K20 (H4K20me2) and H2A ubiquitinated on Lys 15 (H2AK15ub), the latter being a product of RNF168 action on chromatin. 53BP1 binds to nucleosomes minimally as a dimer using its previously characterized methyl-lysine-binding Tudor domain and a carboxy-terminal extension, termed the ubiquitination-dependent recruitment (UDR) motif, which interacts with the epitope formed by H2AK15ub and its surrounding residues on the H2A tail. 53BP1 is therefore a bivalent histone modification reader that recognizes a histone ‘code’ produced by DSB signalling.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Lukas, J., Lukas, C. & Bartek, J. More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nature Cell Biol. 13, 1161–1169 (2011)
Jackson, S. P. & Durocher, D. Regulation of DNA damage responses by ubiquitin and SUMO. Mol. Cell 49, 795–807 (2013)
Nakamura, K. et al. Genetic dissection of vertebrate 53BP1: a major role in non-homologous end joining of DNA double strand breaks. DNA Repair 5, 741–749 (2006)
Bunting, S. F. et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141, 243–254 (2010)
Ward, I. M. et al. 53BP1 is required for class switch recombination. J. Cell Biol. 165, 459–464 (2004)
Manis, J. P. et al. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nature Immunol. 5, 481–487 (2004)
Dimitrova, N., Chen, Y. C., Spector, D. L. & de Lange, T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature 456, 524–528 (2008)
Bouwman, P. et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nature Struct. Mol. Biol. 17, 688–695 (2010)
Bothmer, A. et al. Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol. Cell 42, 319–329 (2011)
Zimmermann, M., Lottersberger, F., Buonomo, S. B., Sfeir, A. & de Lange, T. 53BP1 regulates DSB repair using Rif1 to control 5′ end resection. Science 339, 700–704 (2013)
Escribano-Díaz, C. et al. A cell cycle-dependent regulatory circuit composed of 53BP1–RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell 49, 872–883 (2013)
Di Virgilio, M. et al. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science 339, 711–715 (2013)
Chapman, J. R. et al. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol. Cell 49, 858–871 (2013)
Botuyan, M. V. et al. Structural basis for the methylation state-specific recognition of histone H4–K20 by 53BP1 and Crb2 in DNA repair. Cell 127, 1361–1373 (2006)
Huyen, Y. et al. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 432, 406–411 (2004)
Sanders, S. L. et al. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell 119, 603–614 (2004)
Pei, H. et al. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature 470, 124–128 (2011)
Acs, K. et al. The AAA-ATPase VCP/p97 promotes 53BP1 recruitment by removing L3MBTL1 from DNA double-strand breaks. Nature Struct. Mol. Biol. 18, 1345–1350 (2011)
Mallette, F. A. et al. RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. EMBO J. 31, 1865–1878 (2012)
Nakamura, T. M., Moser, B. A., Du, L. L. & Russell, P. Cooperative control of Crb2 by ATM family and Cdc2 kinases is essential for the DNA damage checkpoint in fission yeast. Mol. Cell. Biol. 25, 10721–10730 (2005)
Zgheib, O., Pataky, K., Brugger, J. & Halazonetis, T. D. An oligomerized 53BP1 tudor domain suffices for recognition of DNA double-strand breaks. Mol. Cell. Biol. 29, 1050–1058 (2009)
Pryde, F. et al. 53BP1 exchanges slowly at the sites of DNA damage and appears to require RNA for its association with chromatin. J. Cell Sci. 118, 2043–2055 (2005)
Gudjonsson, T. et al. TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell 150, 697–709 (2012)
Stewart, G. S. et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 136, 420–434 (2009)
Panier, S. et al. Tandem protein interaction modules organize the ubiquitin-dependent response to DNA double-strand breaks. Mol. Cell 47, 383–395 (2012)
Mattiroli, F. et al. RNF168 ubiquitinates K13–15 on H2A/H2AX to drive DNA damage signaling. Cell 150, 1182–1195 (2012)
Gatti, M. et al. A novel ubiquitin mark at the N-terminal tail of histone H2As targeted by RNF168 ubiquitin ligase. Cell Cycle 11, 2538–2544 (2012)
Wang, H. et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature 431, 873–878 (2004)
Simon, M. D. et al. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell 128, 1003–1012 (2007)
Dikic, I., Wakatsuki, S. & Walters, K. J. Ubiquitin-binding domains—from structures to functions. Nature Rev. Mol. Cell Biol. 10, 659–671 (2009)
Chen, J., Ai, Y., Wang, J., Haracska, L. & Zhuang, Z. Chemically ubiquitylated PCNA as a probe for eukaryotic translesion DNA synthesis. Nature Chem. Biol. 6, 270–272 (2010)
Wu, L. et al. ASH2L regulates ubiquitylation signaling to MLL: trans-regulation of H3 K4 methylation in higher eukaryotes. Mol. Cell 49, 1108–1120 (2013)
Poulsen, M., Lukas, C., Lukas, J., Bekker-Jensen, S. & Mailand, N. Human RNF169 is a negative regulator of the ubiquitin-dependent response to DNA double-strand breaks. J. Cell Biol. 197, 189–199 (2012)
Juang, Y. C. et al. OTUB1 co-opts Lys48-linked ubiquitin recognition to suppress E2 enzyme function. Mol. Cell 45, 384–397 (2012)
Morita, S., Kojima, T. & Kitamura, T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7, 1063–1066 (2000)
Dyer, P. N. et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 375, 23–44 (2004)
Luger, K., Mader, A. W., Richmond, R. K., Sargent, D. F. & Richmond, T. J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260 (1997)
Nakamura, T. M., Du, L. L., Redon, C. & Russell, P. Histone H2A phosphorylation controls Crb2 recruitment at DNA breaks, maintains checkpoint arrest, and influences DNA repair in fission yeast. Mol. Cell. Biol. 24, 6215–6230 (2004)
Bentley, M. L. et al. Recognition of UbcH5c and the nucleosome by the Bmi1/Ring1b ubiquitin ligase complex. EMBO J. 30, 3285–3297 (2011)
Wang, A. C., Grzesiek, S., Tschudin, R., Lodi, P. J. & Bax, A. Sequential backbone assignment of isotopically enriched proteins in D2O by deuterium-decoupled HA(CA)N and HA(CACO)N. J. Biomol. NMR 5, 376–382 (1995)
Acknowledgements
We are grateful to R. Szilard, J. Côté and S. Panier for critically reading the manuscript; to L.-L. Du, A. Nussenzweig, Y. Tong and C. Arrowsmith for plasmids; to M. Cook and A. Rosebrock for centrifugal elutriation; and to B. Sauriol for help with FRAP data analysis. A.F.-T. and A.O. both receive post-doctoral fellowships from the CIHR; C.E.-D. is an Ontario Post-doctoral Fellow; J.K.-L. receives a post-doctoral fellowship from the Leukemia and Lymphoma Society; and C.C.Y.L. and M.-C.L. are post-doctoral fellows of the Canadian Breast Cancer Foundation (Ontario Division). D.D. is the Thomas Kierans Chair in Mechanisms of Cancer Development and a Canada Research Chair (Tier 1) in the Molecular Mechanisms of Genome Integrity. Work in the D.D. laboratory was supported by CIHR grant MOP84297 and grant GL2-01-010 from the Ontario Research Fund.
Author information
Authors and Affiliations
Contributions
A.F.-T. initiated the project, carried out most of the cell biological experiments, found the interaction between ubiquitinated H2A and 53BP1, and contributed to the experimental design and data interpretation. M.D.C. produced the recombinant nucleosomes. A.F.-T. and M.D.C. and carried out the recombinant nucleosome pull-down studies. C.E.-D. examined RIF1 focus formation, the role of the UDR in homologous recombination and helped with some immunofluorescence experiments. A.O. carried out the class switching experiments and the DT40 work. C.C.Y.L. helped with protein binding studies and generated the nucleosome model shown in Supplementary Fig. 12. M.-C.L. helped with the FRAP experiments. J.K.-L. carried out chemical ubiquitination and mass spectrometry to verify histone methylation. H.H. carried out the NMR experiments. S.M.N. carried out mass spectrometry to verify H4K20 methylation. F.S. supervised H.H. D.D. supervised the project and wrote the manuscript with input from the other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-14, Supplementary Text and Data and additional references. (PDF 10429 kb)
Source data
Rights and permissions
About this article
Cite this article
Fradet-Turcotte, A., Canny, M., Escribano-Díaz, C. et al. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature 499, 50–54 (2013). https://doi.org/10.1038/nature12318
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12318
This article is cited by
-
Possible mechanisms and simulation modeling of FLASH radiotherapy
Radiological Physics and Technology (2024)
-
Rif1 Regulates Self-Renewal and Impedes Mesendodermal Differentiation of Mouse Embryonic Stem Cells
Stem Cell Reviews and Reports (2023)
-
Roles for the methyltransferase SETD8 in DNA damage repair
Clinical Epigenetics (2022)
-
DNA damage-induced degradation of Sp1 promotes cellular senescence
GeroScience (2022)
-
Dynamic recruitment of UFM1-specific peptidase 2 to the DNA double-strand breaks regulated by WIP1
Genome Instability & Disease (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.