Abstract
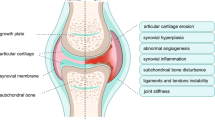
Osteoarthritis (OA), the most common musculoskeletal disorder, is complex, multifaceted, and characterized by degradation of articular cartilage and alterations in other joint tissues. Although some pathogenic pathways have been characterized, current knowledge is incomplete and effective approaches to the prevention or treatment of OA are lacking. Understanding novel molecular mechanisms that are involved in the maintenance and destruction of articular cartilage, including extracellular regulators and intracellular signalling mechanisms in joint cells that control cartilage homeostasis, has the potential to identify new therapeutic targets in OA. MicroRNAs control tissue development and homeostasis by fine-tuning gene expression, with expression patterns specific to tissues and developmental stages, and are increasingly implicated in the pathogenesis of complex diseases such as cancer and cardiovascular disorders. The emergent roles of microRNAs in cartilage homeostasis and OA pathogenesis are summarized in this Review, alongside potential clinical applications.
Key Points
-
Although some pathogenic pathways in osteoarthritis (OA) have been characterized, effective approaches to prevention or treatment of OA are lacking
-
Novel genetic regulators, microRNAs (miRNAs), are involved in the development of the musculoskeletal system and OA pathogenesis through maintenance of articular chondrocytes
-
miRNAs have a role in transmitting the effects of the main risk factors for OA, such as ageing and inflammation, onto cellular homeostasis through their control of multiple target genes
-
Approaches to maintaining or suppressing the expression of key miRNAs in OA pathogenesis have the potential to identify new therapeutic and diagnostic targets
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Helmick, C. G. et al. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States. Part I. Arthritis Rheum. 58, 15–25 (2008).
Lawrence, R. C. et al. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States. Part II. Arthritis Rheum. 58, 26–35 (2008).
Lotz, M. K. & Kraus, V. B. New developments in osteoarthritis. Posttraumatic osteoarthritis: pathogenesis and pharmacological treatment options. Arthritis Res. Ther. 12, 211 (2010).
Hashimoto, M., Nakasa, T., Hikata, T. & Asahara, H. Molecular network of cartilage homeostasis and osteoarthritis. Med. Res. Rev. 28, 464–481 (2008).
Goldring, M. B. & Marcu, K. B. Cartilage homeostasis in health and rheumatic diseases. Arthritis Res. Ther. 11, 224 (2009).
Lotz, M. K. & Caramés, B. Autophagy and cartilage homeostasis mechanisms in joint health, aging and OA. Nat. Rev. Rheumatol. 7, 579–587 (2011).
Farh, K. K. et al. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science 310, 1817–1821 (2005).
Bartel, D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004).
Calin, G. A. & Croce, C. M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 6, 857–866 (2006).
Hafner, M. et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141, 129–141 (2010).
Rao, P. K., Kumar, R. M., Farkhondeh, M., Baskerville, S. & Lodish, H. F. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc. Natl Acad. Sci. USA 103, 8721–8726 (2006).
Taganov, K. D., Boldin, M. P., Chang, K. J. & Baltimore, D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl Acad. Sci. USA 103, 12481–12486 (2006).
Chendrimada, T. P. et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436, 740–744 (2005).
Lee, Y. et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 425, 415–419 (2003).
Gregory, R. I. et al. The Microprocessor complex mediates the genesis of microRNAs. Nature 432, 235–240 (2004).
Denli, A. M., Tops, B. B., Plasterk, R. H., Ketting, R. F. & Hannon, G. J. Processing of primary microRNAs by the Microprocessor complex. Nature 432, 231–235 (2004).
Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 12, 861–874 (2011).
Lewis, B. P., Burge, C. B. & Bartel, D. P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20 (2005).
Bernstein, E. et al. Dicer is essential for mouse development. Nat. Genet. 35, 215–217 (2003).
Kanellopoulou, C. et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 19, 489–501 (2005).
Wang, Y., Medvid, R., Melton, C., Jaenisch, R. & Blelloch, R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 39, 380–385 (2007).
Harfe, B. D., McManus, M. T., Mansfield, J. H., Hornstein, E. & Tabin, C. J. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc. Natl Acad. Sci. USA 102, 10898–10903 (2005).
Kobayashi, T. et al. Dicer-dependent pathways regulate chondrocyte proliferation and differentiation. Proc. Natl Acad. Sci. USA 105, 1949–1954 (2008).
Mizoguchi, F. et al. Osteoclast-specific Dicer gene deficiency suppresses osteoclastic bone resorption. J. Cell. Biochem. 109, 866–875 (2010).
Volinia, S. et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl Acad. Sci. USA 103, 2257–2261 (2006).
Small, E. M. & Olson, E. N. Pervasive roles of microRNAs in cardiovascular biology. Nature 469, 336–342 (2011).
Park, C. Y., Choi, Y. S. & McManus, M. T. Analysis of microRNA knockouts in mice. Hum. Mol. Genet. 19, R169–R175 (2010).
Lanford, R. E. et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 327, 198–201 (2010).
Wienholds, E. et al. MicroRNA expression in zebrafish embryonic development. Science 309, 310–311 (2005).
Eberhart, J. K. et al. MicroRNA Mirn140 modulates Pdgf signaling during palatogenesis. Nat. Genet. 40, 290–298 (2008).
Tuddenham, L. et al. The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett. 580, 4214–4217 (2006).
Miyaki, S. et al. MicroRNA-140 is expressed in differentiated human articular chondrocytes and modulates interleukin-1 responses. Arthritis Rheum. 60, 2723–2730 (2009).
Yamashita, S. et al. L-Sox5 and Sox6 proteins enhance chondrogenic miR-140 microRNA expression by strengthening dimeric Sox9 activity. J. Biol. Chem. 287, 22206–22215 (2012).
Lin, L., Shen, Q., Zhang, C., Chen, L., & Yu, C. Assessment of the profiling microRNA expression of differentiated and dedifferentiated human adult articular chondrocytes. J. Orthop. Res. 29, 1578–1584 (2011).
Miyaki, S. et al. MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes Dev. 24, 1173–1185 (2010).
Nakamura, Y., Inloes, J. B., Katagiri, T. & Kobayashi, T. Chondrocyte-specific microRNA-140 regulates endochondral bone development and targets Dnpep to modulate bone morphogenetic protein signaling. Mol. Cell Biol. 31, 3019–3028 (2011).
Yang, J. et al. MiR-140 is co-expressed with Wwp2-C transcript and activated by Sox9 to target Sp1 in maintaining the chondrocyte proliferation. FEBS Lett. 585, 2992–2997 (2011).
Nicolas, F. E. et al. mRNA expression profiling reveals conserved and non-conserved miR-140 targets. RNA Biol. 8, 607–615 (2011).
Nakamura, Y. et al. Sox9 is upstream of microRNA-140 in cartilage. Appl. Biochem. Biotechnol. 166, 64–71 (2012).
Dudek, K. A., Lafont, J. E., Martinez-Sanchez, A. & Murphy, C. L. Type II collagen expression is regulated by tissue-specific miR-675 in human articular chondrocytes. J. Biol. Chem. 285, 24381–24387 (2010).
Swingler, T. E. et al. The expression and function of microRNAs in chondrogenesis and osteoarthritis. Arthritis Rheum. 64, 1909–1919 (2012).
Martinez-Sanchez, A., Dudek, K. A., & Murphy, C. L. Regulation of human chondrocyte function through direct inhibition of cartilage master regulator SOX9 by microRNA-145 (miRNA-145). J. Biol. Chem. 287, 916–924 (2012).
Xin, M. et al. MicroRNA miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 23, 2166–2178 (2009).
Iliopoulos, D., Malizos, K. N., Oikonomou, P. & Tsezou, A. Integrative microRNA and proteomic approaches identify novel osteoarthritis genes and their collaborative metabolic and inflammatory networks. PLoS One 3, e3740 (2008).
Jones, S. W. et al. The identification of differentially expressed microRNA in osteoarthritic tissue that modulate the production of TNF-α and MMP13. Osteoarthritis Cartilage 17, 464–472 (2009).
Tardif, G., Hum, D., Pelletier, J. P., Duval, N. & Martel-Pelletier, J. Regulation of the IGFBP-5 and MMP-13 genes by the microRNAs miR-140 and miR-27a in human osteoarthritic chondrocytes. BMC Musculoskelet Disord. 10, 148 (2009).
Chen, L. H., Chiou, G. Y., Chen, Y. W., Li, H. Y. & Chiou, S. H. MicroRNA and aging: a novel modulator in regulating the aging network. Ageing Res. Rev. 9 (Suppl. 1), 59–66 (2010).
Liang, R., Bates, D. J. & Wang, E. Epigenetic control of microRNA expression and aging. Curr. Genomics 10, 184–193 (2009).
Gagarina, V. et al. SirT1 enhances survival of human osteoarthritic chondrocytes by repressing protein tyrosine phosphatase 1B and activating the insulin-like growth factor receptor pathway. Arthritis Rheum. 62, 1383–1392 (2010).
Hong, E. H. et al. Ionizing radiation induces cellular senescence of articular chondrocytes via negative regulation of SIRT1 by p38 kinase. J. Biol. Chem. 285, 1283–1295 (2010).
Takayama, K. et al. SIRT1 regulation of apoptosis of human chondrocytes. Arthritis Rheum. 60, 2731–2740 (2009).
Fujita, N. et al. Potential involvement of SIRT1 in the pathogenesis of osteoarthritis through the modulation of chondrocyte gene expressions. J. Orthop. Res. 29, 511–515 (2011).
Dvir-Ginzberg, M., Gagarina, V., Lee, E. J. & Hall, D. J. Regulation of cartilage-specific gene expression in human chondrocytes by SirT1 and nicotinamide phosphoribosyltransferase. J. Biol. Chem. 283, 36300–36310 (2008).
Saunders, L. R. et al. miRNAs regulate SIRT1 expression during mouse embryonic stem cell differentiation and in adult mouse tissues. Aging (Albany NY) 2, 415–431 (2010).
Schonrock, N., Humphreys, D. T., Preiss, T. & Götz, J. Target gene repression mediated by miRNAs miR-181c and miR-9 both of which are down-regulated by amyloid-β. J. Mol. Neurosci. 46, 324–335 (2012).
Xu, D. et al. miR-22 represses cancer progression by inducing cellular senescence. J. Cell. Biol. 193, 409–424 (2011).
Jazbutyte, V. et al. MicroRNA-22 increases senescence and activates cardiac fibroblasts in the aging heart. Age (Dordr.) (in press).
Abouheif, M. M. et al. Silencing microRNA-34a inhibits chondrocytes apoptosis in a rat osteoarthritis model in vitro. Rheumatology (Oxford) 49, 2054–2060 (2010).
Dunn, W., DuRaine, G. & Reddi, A. H. Profiling microRNA expression in bovine articular cartilage and implications for mechanotransduction. Arthritis Rheum. 60, 2333–2339 (2009).
Ceribelli, A., Nahid, M. A., Satoh, M. & Chan, E. K. MicroRNAs in rheumatoid arthritis. FEBS Lett. 585, 3667–3674 (2011).
Stanczyk, J. et al. Altered expression of microRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 58, 1001–1009 (2008).
Nakasa, T. et al. Expression of microRNA-146 in rheumatoid arthritis synovial tissue. Arthritis Rheum. 58, 1284–1292 (2008).
Yamasaki, K. et al. Expression of microRNA-146a in osteoarthritis cartilage. Arthritis Rheum. 60, 1035–1041 (2009).
Akhtar, N. et al. MicroRNA-27b regulates the expression of matrix metalloproteinase 13 in human osteoarthritis chondrocytes. Arthritis Rheum. 62, 1361–1371 (2010).
Li, X. et al. MicroRNA-146a is linked to pain-related pathophysiology of osteoarthritis. Gene 480, 34–41 (2011).
Suri, S. & Walsh, D. A. Osteochondral alterations in osteoarthritis. Bone 51, 204–211 (2012).
Pauley, K. M. et al. Upregulated miR-146a expression in peripheral blood mononuclear cells from rheumatoid arthritis patients. Arthritis Res. Ther. 10, R101 (2008).
Okuhara, A. et al. Change in microRNA expression in peripheral mononuclear cells according to the progression of osteoarthritis. Mod. Rheumatol. 22, 446–457 (2012).
Nakasa, T., Shibuya, H., Nagata, Y., Niimoto, T. & Ochi, M. The inhibitory effect of microRNA-146a expression on bone destruction in collagen-induced arthritis. Arthritis Rheum. 63, 1582–1590 (2011).
Sugatani, T. & Hruska, K. A. MicroRNA-223 is a key factor in osteoclast differentiation. J. Cell Biochem. 101, 996–999 (2007).
Sugatani, T. & Hruska, K. A. Impaired micro-RNA pathways diminish osteoclast differentiation and function. J. Biol. Chem. 284, 4667–4678 (2009).
Mizuno, Y. et al. miR-210 promotes osteoblastic differentiation through inhibition of AcvR1b. FEBS Lett. 583, 2263–2268 (2009).
Fasanaro, P. et al. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J. Biol. Chem. 283, 15878–15883 (2008).
Li, H. et al. A novel microRNA targeting HDAC5 regulates osteoblast differentiation in mice and contributes to primary osteoporosis in humans. J. Clin. Invest. 119, 3666–3677 (2009).
Hu, R. et al. A Runx/miR-3960/miR-2861 regulatory feedback loop during mouse osteoblast differentiation. J. Biol. Chem. 286, 12328–12339 (2011).
Echtermeyer, F. et al. Syndecan-4 regulates ADAMTS-5 activation and cartilage breakdown in osteoarthritis. Nat. Med. 15, 1072–1076 (2009).
Glasson, S. S. et al. Deletion of active ADAMTS5 prevents cartilage degradation in a murine model of osteoarthritis. Nature 434, 644–648 (2005).
Stanton, H. et al. ADAMTS5 is the major aggrecanase in mouse cartilage in vivo and in vitro. Nature 434, 648–652 (2005).
Saito, T. et al. Transcriptional regulation of endochondral ossification by HIF-2α during skeletal growth and osteoarthritis development. Nat. Med. 16, 678–686 (2010).
Yang, S. et al. Hypoxia-inducible factor-2α is a catabolic regulator of osteoarthritic cartilage destruction. Nat. Med. 16, 687–693 (2010).
Otsuki, S. et al. Extracellular sulfatases support cartilage homeostasis by regulating BMP and FGF signaling pathways. Proc. Natl Acad. Sci. USA 107, 10202–10207 (2010).
Taniguchi, N. et al. Aging-related loss of the chromatin protein HMGB2 in articular cartilage is linked to reduced cellularity and osteoarthritis. Proc. Natl Acad. Sci. USA 106, 1181–1186 (2009).
Hellio Le Graverand-Gastineau, M. P. OA clinical trials: current targets and trials for OA. Choosing molecular targets: what have we learned and where we are headed? Osteoarthritis Cartilage 17, 1393–1401 (2009).
Thirunavukkarasu, K., Pei, Y. & Wei, T. Characterization of the human ADAMTS-5 (aggrecanase-2) gene promoter. Mol. Biol. Rep. 34, 225–231 (2007).
Lin, A. C. et al. Modulating hedgehog signaling can attenuate the severity of osteoarthritis. Nat. Med. 15, 1421–1425 (2009).
Saito, T. & Kawaguchi, H. HIF-2α as a possible therapeutic target of osteoarthritis. Osteoarthritis Cartilage 18, 1552–1556 (2010).
Alcaraz, M. J., Megías, J., García-Arnandis, I., Clérigues V & Guillén, M. I. New molecular targets for the treatment of osteoarthritis. Biochem. Pharmacol. 80, 13–21 (2010).
Eguchi, A. et al. Efficient siRNA delivery into primary cells by a peptide transduction domain-dsRNA binding domain fusion protein. Nat. Biotechnol. 27, 567–571 (2009).
Nagata, Y. et al. Induction of apoptosis in the synovium of mice with autoantibody-mediated arthritis by the intraarticular injection of double-stranded microRNA-15a. Arthritis Rheum. 60, 2677–2683 (2009).
Mitchell, P. S. et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl Acad. Sci. USA 105, 10513–10518 (2008).
Kosaka, N., Iguchi, H. & Ochiya, T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 101, 2087–2092 (2010).
Wittmann, J. & Jäck, H. M. Serum microRNAs as powerful cancer biomarkers. Biochim. Biophys. Acta 1806, 200–207 (2010).
Murata, K. et al. Plasma and synovial fluid microRNAs as potential biomarkers of rheumatoid arthritis and osteoarthritis. Arthritis Res. Ther. 12, R86 (2010).
Kosaka, N., Izumi, H., Sekine, K. & Ochiya, T. MicroRNA as a new immune-regulatory agent in breast milk. Silence 1, 7 (2010).
Alvarez-Erviti, L. et al. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 29, 341–345 (2011).
Acknowledgements
This work is partly supported by the NIH (USA; grant numbers AR050631 and AR056120), the Arthritis National Research Foundation (USA), Health and Labour Sciences Research Grants (Ministry of Health, Labour and Welfare, Japan), Grants-in-Aid for Scientific Research (Ministry of Education, Culture, Sports, Science and Technology, Japan), the National Center for Child Health and Development (Japan; 20A-3) and CREST (Japan Science and Technology Agency).
Author information
Authors and Affiliations
Contributions
Both authors contributed equally to researching data for the article, discussing its content, writing and review/editing of the manuscript before publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Miyaki, S., Asahara, H. Macro view of microRNA function in osteoarthritis. Nat Rev Rheumatol 8, 543–552 (2012). https://doi.org/10.1038/nrrheum.2012.128
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrrheum.2012.128
This article is cited by
-
Mechanical overloading-induced miR-325-3p reduction promoted chondrocyte senescence and exacerbated facet joint degeneration
Arthritis Research & Therapy (2023)
-
Circulating miR-146b and miR-27b are efficient biomarkers for early diagnosis of Equidae osteoarthritis
Scientific Reports (2023)
-
Exploration of microRNA-106b-5p as a therapeutic target in intervertebral disc degeneration: a preclinical study
Apoptosis (2023)
-
Focusing on the hypoxia-inducible factor pathway: role, regulation, and therapy for osteoarthritis
European Journal of Medical Research (2022)
-
MiR-99a alleviates apoptosis and extracellular matrix degradation in experimentally induced spine osteoarthritis by targeting FZD8
BMC Musculoskeletal Disorders (2022)