Abstract
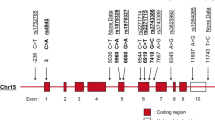
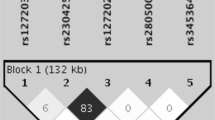
Systemic lupus erythematosus (SLE) is a multisystem complex autoimmune disease of uncertain etiology (OMIM 152700). Over recent years a genetic component to SLE susceptibility has been established1,2,3. Recent successes with association studies in SLE have identified genes including IRF5 (refs. 4,5) and FCGR3B6. Two tumor necrosis factor (TNF) superfamily members located within intervals showing genetic linkage with SLE are TNFSF4 (also known as OX40L; 1q25), which is expressed on activated antigen-presenting cells (APCs)7,8 and vascular endothelial cells9, and also its unique receptor, TNFRSF4 (also known as OX40; 1p36), which is primarily expressed on activated CD4+ T cells10. TNFSF4 produces a potent co-stimulatory signal for activated CD4+ T cells after engagement of TNFRSF4 (ref. 11). Using both a family-based and a case-control study design, we show that the upstream region of TNFSF4 contains a single risk haplotype for SLE, which is correlated with increased expression of both cell-surface TNFSF4 and the TNFSF4 transcript. We hypothesize that increased expression of TNFSF4 predisposes to SLE either by quantitatively augmenting T cell–APC interaction or by influencing the functional consequences of T cell activation via TNFRSF4.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Accession codes
References
Vyse, T.J. & Todd, J.A. Genetic analysis of autoimmune disease. Cell 85, 311–318 (1996).
Deapen, D. et al. A revised estimate of twin concordance in systemic lupus erythematosus. Arthritis Rheum. 35, 311–318 (1992).
Forabosco, P. et al. Meta-analysis of genome-wide linkage studies of systemic lupus erythematosus. Genes Immun. 7, 609–614 (2006).
Graham, R.R. et al. A common haplotype of interferon regulatory factor 5 (IRF5) regulates splicing and expression and is associated with increased risk of systemic lupus erythematosus. Nat. Genet. 38, 550–555 (2006).
Cunninghame Graham, D.S. et al. Association of IRF5 in UK SLE families identifies a variant involved in polyadenylation. Hum. Mol. Genet. 16, 579–591 (2007).
Fanciulli, M. et al. FCGR3B copy number variation is associated with susceptibility to systemic, but not organ-specific, autoimmunity. Nat. Genet. 39, 721–723 (2007).
Ohshima, Y. et al. Expression and function of OX40 ligand on human dendritic cells. J. Immunol. 159, 3838–3848 (1997).
Godfrey, W.R., Fagnoni, F.F., Harara, M.A., Buck, D. & Engleman, E.G. Identification of a human OX-40 ligand, a costimulator of CD4+ T cells with homology to tumor necrosis factor. J. Exp. Med. 180, 757–762 (1994).
Imura, A. et al. The human OX40/gp34 system directly mediates adhesion of activated T cells to vascular endothelial cells. J. Exp. Med. 183, 2185–2195 (1996).
Latza, U. et al. The human OX40 homolog: cDNA structure, expression and chromosomal assignment of the ACT35 antigen. Eur. J. Immunol. 24, 677–683 (1994).
Gramaglia, I., Weinberg, A.D., Lemon, M. & Croft, M. Ox-40 ligand: a potent costimulatory molecule for sustaining primary CD4 T cell responses. J. Immunol. 161, 6510–6517 (1998).
Ikushima, S. et al. Pivotal role for the NFIL3/E4BP4 transcription factor in interleukin 3-mediated survival of pro-B lymphocytes. Proc. Natl. Acad. Sci. USA 94, 2609–2614 (1997).
Stuber, E., Neurath, M., Calderhead, D., Fell, H.P. & Strober, W. Cross-linking of OX40 ligand, a member of the TNF/NGF cytokine family, induces proliferation and differentiation in murine splenic B cells. Immunity 2, 507–521 (1995).
Ito, T. et al. OX40 ligand shuts down IL-10-producing regulatory T cells. Proc. Natl. Acad. Sci. USA 103, 13138–13143 (2006).
Wang, X. et al. Positional identification of TNFSF4, encoding OX40 ligand, as a gene that influences atherosclerosis susceptibility. Nat. Genet. 37, 365–372 (2005).
Graham, D.S., Wong, A.K., McHugh, N.J., Whittaker, J.C. & Vyse, T.J. Evidence for unique association signals in SLE at the CD28-CTLA4-ICOS locus in a family-based study. Hum. Mol. Genet. 15, 3195–3205 (2006).
Tan, E.M. et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 25, 1271–1277 (1982).
Boyum, A. Separation of leukocytes from blood and bone marrow. Introduction. Scand. J. Clin. Lab. Invest. Suppl. 97, 7 (1968).
Storm, N., Darnhofer-Patel, B., van den Boom, D. & Rodi, C.P. MALDI-TOF mass spectrometry-based SNP genotyping. Methods Mol. Biol. 212, 241–262 (2003).
Gabriel, S.B. et al. The structure of haplotype blocks in the human genome. Science 296, 2225–2229 (2002).
Lewontin, R.C. The interaction of selection and linkage. I. General considerations; heterotic models. Genetics 49, 49–67 (1964).
Hill, W.G. & Weir, B.S. Maximum-likelihood estimation of gene location by linkage disequilibrium. Am. J. Hum. Genet. 54, 705–714 (1994).
Devlin, B. & Risch, N. A comparison of linkage disequilibrium measures for fine-scale mapping. Genomics 29, 311–322 (1995).
Kruglyak, L., Daly, M.J., Reeve-Daly, M.P. & Lander, E.S. Parametric and nonparametric linkage analysis: a unified multipoint approach. Am. J. Hum. Genet. 58, 1347–1363 (1996).
Kruglyak, L. & Lander, E.S. Faster multipoint linkage analysis using Fourier transforms. J. Comput. Biol. 5, 1–7 (1998).
Fisher, R.A. Statistical Methods for Research Workers (Hafner, New York, 1954).
Clayton, D. A generalization of the transmission/disequilibrium test for uncertain-haplotype transmission. Am. J. Hum. Genet. 65, 1170–1177 (1999).
Stephens, M., Smith, N.J. & Donnelly, P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 68, 978–989 (2001).
Acknowledgements
This work was funded by the Wellcome Trust through a Senior Fellowship awarded to T.J.V. We acknowledge the work of P. Spencer in recruiting individuals and families into the study, and we would like to thank our clinical colleagues for helping us recruit study participants. Our thanks and appreciation is extended to all the study participants and their relatives for generously donating blood samples, and to all the general practitioners and practice nurses for collecting them. J.D.R. is funded by grants from the National Institutes of Allergy and Infectious Diseases (AI065687; AI067152) and from the National Institute of Diabetes and Digestive and Kidney Diseases (DK064869; DK062432). We appreciate the contribution to the genotyping from the Broad Institute Center for Genotyping and Analysis, which is supported by grant U54 RR020278-01 from the National Center for Research Resources. We gratefully acknowledge the work of D. Smyth at the Juvenile Diabetes Research Foundation/Wellcome Trust Diabetes and Inflammation Laboratory for genotyping five variants from the diabetes collection. We also thank H. Stevens and the Diabetes and Inflammation Laboratory DNA team for sending EBV-transformed lymphoblastoid cell lines. We acknowledge use of DNA from the British 1958 Birth Cohort collection (D. Strachan, S. Ring, W. McArdle and M. Pembrey), funded by the Medical Research Council grant G0000934 and Wellcome Trust grant 068545/Z/02. We thank J. Todd for LCL lymphocytes and for critical reading of this manuscript.
Author information
Authors and Affiliations
Contributions
D.S.C.G., T.J.V., H.M. and R.R.G. wrote the manuscript, A.K.W. managed the database, T.J.V. collected the UK samples, T.W.B., P.M.G. and K.L.M. provided access to the Minnesota samples, D.S.C.G. and J.D.R. performed the UK genotyping, R.R.G. and D.A. carried out the Minnesota genotyping, H.M. did the expression analysis, A.K.W. collected the lymphocytes, D.S.C.G., J.C.W. and J.D.R. did the data analysis in the UK samples and R.R.G. contributed to the analysis of the Minnesota samples.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–6 and Supplementary Figures 1 and 2 (PDF 199 kb)
Rights and permissions
About this article
Cite this article
Graham, D., Graham, R., Manku, H. et al. Polymorphism at the TNF superfamily gene TNFSF4 confers susceptibility to systemic lupus erythematosus. Nat Genet 40, 83–89 (2008). https://doi.org/10.1038/ng.2007.47
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2007.47
This article is cited by
-
Immunologic and Genetic Contributors to CD46-Dependent Immune Dysregulation
Journal of Clinical Immunology (2023)
-
Association of TNFSF4 polymorphisms with systemic lupus erythematosus: a meta-analysis
Advances in Rheumatology (2021)
-
TNFSF4 is a risk factor to systemic lupus erythematosus in a Latin American population
Clinical Rheumatology (2021)
-
The haplotypes of various TNF related genes associated with scleritis in Chinese Han
Human Genomics (2020)
-
The Post-GWAS Era: How to Validate the Contribution of Gene Variants in Lupus
Current Rheumatology Reports (2019)