Abstract
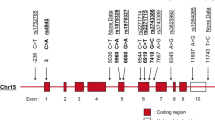
Genetic factors influence susceptibility to systemic lupus erythematosus (SLE). A recent family-based analysis in Caucasian and Chinese populations provided evidence for association of single-nucleotide polymorphisms (SNPs) in the complement receptor 2 (CR2/CD21) gene with SLE. Here we confirmed this result in a case–control analysis of an independent European-derived population including 2084 patients with SLE and 2853 healthy controls. A haplotype formed by the minor alleles of three CR2 SNPs (rs1048971, rs17615, rs4308977) showed significant association with decreased risk of SLE (30.4% in cases vs 32.6% in controls, P=0.016, OR=0.90 (0.82–0.98)). Two of these SNPs are in exon 10, directly 5′ of an alternatively spliced exon preferentially expressed in follicular dendritic cells (FDC), and the third is in the alternatively spliced exon. Effects of these SNPs and a fourth SNP in exon 11 (rs17616) on alternative splicing were evaluated. We found that the minor alleles of these SNPs decreased splicing efficiency of exon 11 both in vitro and ex vivo. These findings further implicate CR2 in the pathogenesis of SLE and suggest that CR2 variants alter the maintenance of tolerance and autoantibody production in the secondary lymphoid tissues where B cells and FDCs interact.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 digital issues and online access to articles
$119.00 per year
only $19.83 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Tsao BP . Update on human systemic lupus erythematosus genetics. Curr Opin Rheumatol 2004; 16: 513–521.
Wu H, Boackle SA, Hanvivadhanakul P, Ulgiati D, Grossman JM, Lee Y et al. Association of a common complement receptor 2 haplotype with increased risk of systemic lupus erythematosus. Proc Natl Acad Sci USA 2007; 104: 3961–3966.
Boackle SA, Holers VM, Chen X, Szakonyi G, Karp DR, Wakeland EK et al. Cr2, a candidate gene in the murine Sle1c lupus susceptibility locus, encodes a dysfunctional protein. Immunity 2001; 15: 775–785.
Miyagawa H, Yamai M, Sakaguchi D, Kiyohara C, Tsukamoto H, Kimoto Y et al. Association of polymorphisms in complement component C3 gene with susceptibility to systemic lupus erythematosus. Rheumatology (Oxford) 2008; 47: 158–164.
Weis JJ, Toothaker LE, Smith JA, Weis JH, Fearon DT . Structure of the human B lymphocyte receptor for C3d and the Epstein-Barr virus and relatedness to other members of the family of C3/C4 proteins. JExpMed 1988; 167: 1047–1066.
Moore MD, Cooper NR, Tack BF, Nemerow GR . Molecular cloning of the cDNA encoding the Epstein-Barr virus/C3d receptor (complement receptor type 2) of human B lymphocytes. Proc Natl Acad Sci USA 1987; 84: 9194–9198.
Fujisaku A, Harley JB, Frank MB, Gruner BA, Frazier B, Holers VM . Genomic organization and polymorphisms of the human C3d/Epstein-Barr virus receptor. J Biol Chem 1989; 264: 2118–2125.
Liu Y-J, Xu J, de Bouteiller O, Parham CL, Grouard G, Djossou O et al. Follicular dendritic cells specifically express the long CR2/CD21 isoform. J Exp Med 1997; 185: 165–170.
Fingeroth JD, Weis JJ, Tedder TF, Strominger JL, Biro PA, Fearon DT . Epstein-Barr virus receptor of human B lymphocytes is the C3d receptor CR2. Proc Natl Acad Sci USA 1984; 81: 4510–4514.
Aubry J-P, Pochon S, Graber P, Jansen KU, Bonnefoy J-Y . CD21 is a ligand for CD23 and regulates IgE production. Nature 1992; 358: 505–507.
Delcayre AX, Salas F, Mathur S, Kovats K, Lotz M, Lernhardt W . Epstein Barr virus/complement C3d receptor is an interferon alpha receptor. EMBO J 1991; 10: 919–926.
Vereshchagina LA, Tolnay M, Tsokos GC . Multiple transcriptional factors regulate the inducible expression of the human complement receptor 2 promoter. J Immunol 2001; 166: 6156–6163.
Tolnay M, Vereshchagina LA, Tsokos GC . NF-kappaB regulates the expression of the human complement receptor 2 gene. J Immunol 2002; 169: 6236–6243.
Ulgiati D, Holers VM . CR2/CD21 proximal promoter activity is critically dependent on a cell type-specific repressor. J Immunol 2001; 167: 6912–6919.
Ulgiati D, Pham C, Holers VM . Functional analysis of the human complement receptor 2 (CR2/CD21) promoter: characterization of basal transcriptional mechanisms. J Immunol 2002; 168: 6279–6285.
Makar KW, Pham CTN, Dehoff MH, O'Connor SM, Jacobi SM, Holers VM . An intronic silencer regulates B lymphocyte cell- and stage-specific expression of the human complement receptor type 2 (CR2, CD21) gene. J Immunol 1998; 160: 1268–1278.
Makar KW, Ulgiati D, Hagman J, Holers VM . A site in the complement receptor 2 (CR2/CD21) silencer is necessary for lineage specific transcriptional regulation. Int Immunol 2001; 13: 657–664.
Holers VM . Complement receptors and the shaping of the natural antibody repertoire. Springer Semin Immun 2005; 26: 405–423.
Hochberg MC . Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 1997; 40: 1725.
Auch D, Reth M . Exon trap cloning: using PCR to rapidly detect and clone exons from genomic DNA fragments. Nucleic Acids Res 1990; 18: 6743–6744.
Kim H-S, Zhang X, Choi YS . Activation and proliferation of follicular dendritic cell-like cells by activated T lymphocytes. J Immunol 1994; 153: 2951–2961.
Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C et al. Alternative isoform regulation in human tissue transcriptomes. Nature 2008; 456: 470–476.
Castle JC, Zhang C, Shah JK, Kulharni AV, Kalsotra A, Cooper TA et al. Expression of 24 426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat Genet 2008; 40: 1416–1425.
Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ . Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 2008; 40: 1413–1415.
Modrek B, Resch A, Grasso C, Lee C . Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res 2001; 29: 2850–2859.
Kwan T, Benovoy D, Dias C, Gurd S, Provencher C, Beaulieu P et al. Genome-wide analysis of transcript isoform variation in humans. Nat Genet 2008; 40: 225–231.
Wang G-S, Cooper TA . Splicing in disease: disruption of the splicing code and the decoding machinery. Nature Rev Genet 2007; 8: 749–761.
Hertel KJ . Combinatorial control of exon recognition. J Biol Chem 2008; 283: 1211–1215.
Hiller M, Zhang Z, Backofen R, Stamm S . Pre-mRNA secondary structures influence exon recognition. PLoS Genetics 2007; 3: 2147–2155.
Kornblihtt AR . Chromatin, transcript elongation and alternative splicing. Nat Struct Mol Biol 2006; 13: 5–7.
Illges H, Braun M, Peter HH, Melchers I . Analysis of the human CD21 transcription unit reveals differential splicing of exon 11 in mature transcripts and excludes alternative splicing as the mechanism causing solubilization of CR2. Mol Immunol 1997; 34: 683–693.
Wu X, Jiang N, Deppong C, Singh J, Dolecki G, Mao D et al. A role for the Cr2 gene in modifying autoantibody production in systemic lupus erythematosus. J Immunol 2002; 169: 1587–1592.
Prodeus AP, Georg S, Shen L-M, Pozdnyakova OO, Chu L, Alicot EM et al. A critical role for complement in the maintenance of self-tolerance. Immunity 1998; 9: 721–731.
Wilson JG, Ratnoff WD, Schur PH, Fearon DT . Decreased expression of the C3b/C4b receptor (CR1) and the C3d receptor (CR2) on B lymphocytes and of CR1 on neutrophils of patients with systemic lupus erythematosus. Arth Rheum 1986; 29: 739–747.
Levy E, Ambrus J, Kahl L, Molina H, Tung K, Holers VM . T lymphocyte expression of complement receptor 2 (CR2/CD21): a role in adhesive cell-cell interactions and dysregulation in a patient with systemic lupus erythematosus (SLE). Clin Exp Immunol 1992; 90: 235–244.
Marquart HV, Svendsen A, Rasmussen JM, Nielsen CH, Junker P, Svehag S-E et al. Complement receptor expression and activation of the complement cascade on B lymphocytes from patients with systemic lupus erythematosus (SLE). Clin Exp Immunol 1995; 101: 60–65.
Matsumoto AK, Martin DR, Carter RH, Klickstein LB, Ahearn JM, Fearon DT . Functional dissection of the CD21/CD19/TAPA-1/Leu-13 complex of B lymphocytes. J Exp Med 1993; 178: 1407–1417.
Steiner B, Truninger K, Sanz J, Schaller A, Gallati S . The role of common single-nucleotide polymorphisms on exon 9 and exon 12 skipping in nonmutated CFTR alleles. Hum Mutat 2004; 24: 120–129.
Cornall RJ, Cyster JG, Hibbs ML, Dunn AR, Otipoby KL, Clark EA et al. Polygenic autoimmune traits: Lyn, CD22, and SHP-1 are limiting elements of a biochemical pathway regulating BCR signaling and selection. Immunity 1998; 8: 497–508.
Morel L, Croker BP, Blenman KR, Mohan C, Huang G, Gilkeson G et al. Genetic reconstitution of systemic lupus erythematosus immunopathology with polycongenic murine strains. Proc Natl Acad Sci USA 2000; 97: 6670–6675.
Giles BM, Tchepeleva SN, Kachinski JJ, Ruff K, Croker BP, Morel L et al. Augmentation of NZB autoimmune phenotypes by the NZW-derived Sle1c lupus susceptibility interval. J Immunol 2007; 178: 4667–4675.
The International Consortium for Systemic Lupus Erythematosus Genetics (SLEGEN), Harley JB, Alarcón-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet 2008; 40: 204–210.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575.
Kazeem GR, Farrall M . Integrating case-control and TDT studies. Ann Hum Genet 2005; 69: 329–335.
Acknowledgements
We thank Dr Raul Torres (National Jewish Health and University of Colorado Denver School of Medicine) for providing the pL53In vector, Dr Yong Choi (Ochsner Clinic Foundation) for providing the HK FDC line, and Carissa Homme and Lauren Kuhlman (University of Colorado Denver School of Medicine, Aurora, CO, USA) for technical assistance. We also thank Dr Peter Gregersen (Feinstein Institute for Medical Research, Manhassat, NY, USA) for contributing DNA samples from control subjects to LLAS, and the Wake Forest University Health Sciences Center for Public Health Genomics for assistance with the principal component analysis of the genotyping data from LLAS1. The pL53In constructs were sequenced by the University of Colorado Cancer Center DNA Sequencing and Analysis Core (http://loki.uchsc.edu), which is supported by the NIH/NCI Cancer Core Support Grant (P30 CA046934). The quality of the RNA samples prepared from healthy human subjects was evaluated in the University of Colorado Cancer Center Microarray Core. Quantitative RT-PCR was carried out in the DERC Molecular Biology Core Facility, which is supported by National Institutes of Health (NIH) grant P30 DK57516. Other support for this work included grants from the American College of Rheumatology Research and Education Foundation, the Alliance for Lupus Research, the US Department of Veterans Affairs, Kirkland Scholar/Hospital for Special Surgery and Rheuminations, and NIH (NIAID-DAIT-BAA-05-11, N01 AR12253, N01 AR62277, P01 AR049084, P20 RR015577, P20 RR020143, P30 AR053483, P50 AR48940, R01 DE015223, R01 AI31584, R01 AI070983R01 AR42460, and R37 AI24717).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Douglas, K., Windels, D., Zhao, J. et al. Complement receptor 2 polymorphisms associated with systemic lupus erythematosus modulate alternative splicing. Genes Immun 10, 457–469 (2009). https://doi.org/10.1038/gene.2009.27
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gene.2009.27
Keywords
This article is cited by
-
Exploring the etiopathogenesis of systemic lupus erythematosus: a genetic perspective
Immunogenetics (2019)
-
Complement gene variants in relation to autoantibodies to beta cell specific antigens and type 1 diabetes in the TEDDY Study
Scientific Reports (2016)
-
Association of complement receptor 2 polymorphisms with innate resistance to HIV-1 infection
Genes & Immunity (2015)
-
Linking complement and anti-dsDNA antibodies in the pathogenesis of systemic lupus erythematosus
Immunologic Research (2013)
-
Genetics and Epigenetics of Systemic Lupus Erythematosus
Current Rheumatology Reports (2013)